Shin Woong-hee of Chemical Education
Lab of Computer-Aided Drug Discovery
Our lab aims to develop computational tools for computer-aided drug discovery (CADD) and to apply them to a real-world system. As the power of a computer increases, the CADD technique gets more interest. It can find and design drug candidates for drug targets, especially proteins. Thus, it helps medicinal chemists by reducing money and labor costs in the drug discovery process.
Aim 1. Develop CADD tools
1.1 Predict binding affinity from a given protein-ligand complex
Binding free energy is a key aspect of a drug design. A stronger binder means a small concentration could treat a target disease. Also, predicting an accurate binding affinity can help to get a precise binding pose, which can get a clue for drug design. Collaborating with Arontier co., we developed an algorithm called AK-Score1 that predicts a binding affinity from a given 3D protein-ligand complex structure by deep learning.
1.2 Investigate protein-ligand interaction using molecular surface
Most molecular interaction occurs at the surface of molecules. However, traditional CADD tools employ an atomic-based model to describe a protein-ligand interaction since it is hard to get molecular surfaces from the atomic model. Even though they successfully applied to predict some drugs, they have two major hurdles: (1) dealing with the flexibility of biomolecules and (2) computational time to calculate atomic distance. To solve the issues, we recently developed a virtual screening program called PL-PatchSurfer2,3 computes a protein-ligand complementarity using molecular surfaces. With the help of Zernike descriptors, PL-PatchSurfer performed best among benchmarked virtual screening programs and showed 20 times faster computation time. In the following project, we are working on combining molecular surface with deep learning to predict a protein-ligand interaction.
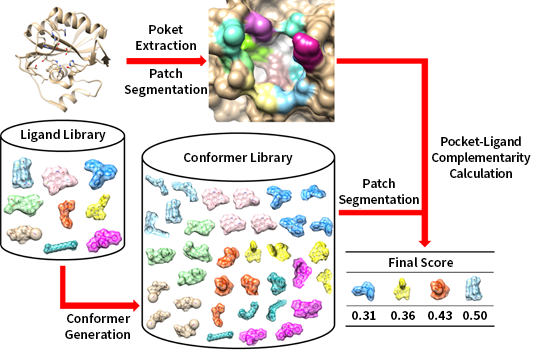
Poket Extraction,Patch Segmentation
Ligand Library, ConFormer Generation
Conformer Library
Patch Segmentation
Pocket-Ligand Complementarity Calculation
Final Score : 0.31, 0.36, 0.43, 0.50
Aim 2. Apply to the real-world problems
Not only for developing the CADD tools, but we also do co-work with wet-lab experimentalists from around the world to improve our program and solve the real problem.
2.1 Identify GLI1 inhibitor as an anti-cancer drug
GLI1 is a transcription factor, located at the end of the Hedgehog pathway. The protein is overexpressed in cancer cells such as glioblastoma, medulloblastoma, and basal cell carcinoma. Most anti-cancer drugs targeting the Hedgehog pathway target smoothened homologs (SMO), a G-Protein-Coupled-Receptor. However, the drugs are reported inactive when a mutation occurs. Thus, targeting GLI1 is more promising than SMO. The long-term goal of this project is to identify novel GLI1 inhibitors to overcome anti-SMO resistance by virtual screening and in vitro experiments.
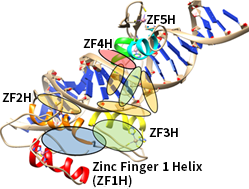
Zinc Finger 1 Helix(ZF1H), ZF2H, ZF3H, ZF4H, ZF5H
2.2 Design anti-cancer drugs by re-invigorate exhausted T cells
Immune checkpoint receptors such as PD-1 and CLTA-4 are highly expressed when the T cells lost their activities. Inhibiting these receptors re-activates T cells, so they can attack cancer cells. Thus, re-invigorating T cells has been highlighted as a new concept for anti-cancer recently. The goals of the project are to discover small compounds and design new protein drugs targeting the immune checkpoint proteins to re-activate T cells.
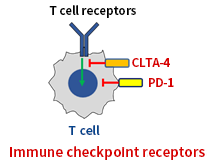
T cell receptors
CLTA-4, PD-1
T cell
Immune checkpoint receptors
- 1Y Kwon, J Ko, WH Shin, and J Lee. AK-Score: Accurate Protein-Ligand Binding Affinity Prediction Using an Ensemble of 3D-Convolutional Neural Networks. Int. J. Mol. Sci. 2020, 21, 8424.
- 2WH Shin, CW Christoffer, J Wang, and D Kihara. PL-PatchSurfer2: Improved Virtual Screening Method Using Local Surface Matching that is Tolerant to Target and Ligand Surface Variation. J. Chem. Inf. Model. 2016, 56, 1676.
- 3WH Shin and D Kihara. Predicting binding poses and affinity ranking in D3R Grand Challenge using PL-PatchSurfer2.0. J. Comput.-Aided Mol. Des. 2019, 33, 1083.
